An Open Source software designed for the study of molecular dynamics on Linux OSes. #Study proteins #Study macromolecules #NMR relaxation data #Nmr-relax #Study #Proteins
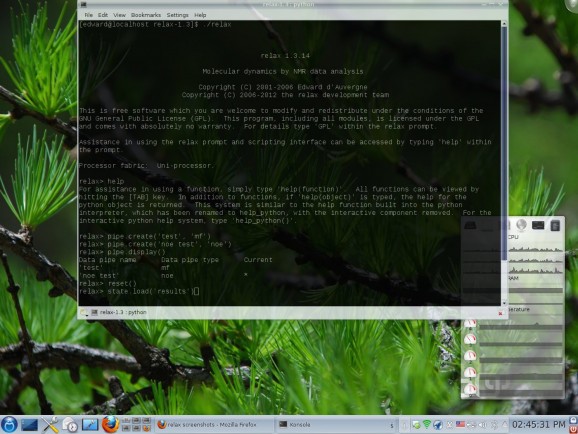
relax is an open source and freely distributed software project that has been designed for studying molecular dynamics by analyzing experimental NMR data, supporting organic molecules, RNA, proteins, sugars, DNA, and many other biomolecules.
relax supports several NMR theories, implements various data analysis tools as modular components, and it can interface with other programs, such as Dasha and Modelfree. It supports a wide range of NMR theories, incorporates several data analysis tools, allows users to visualize data, as well as to interact with other programs.
Despite the fact that the application comes with both a CLI (Command-line Interface) and GUI (Graphical User Interface) front-end, several third-party graphical user interfaces (GUIs) exist for relax.
Among the supported types of analyzes, relax can handle relaxation dispersion, consistency testing of multiple field NMR (Nuclear Magnetic Resonance) relaxation data, N-state model and frame order, model-free analysis, R1 and R2, NOE, RSDM (Reduced Spectral Density Mapping), as well as stereochemistry investigations.
In order to automate the data analysis process, it is possible to create very complex scripts by using building blocks. For this, the developers provide various sample scripts that will help you to understand the script construction and to more easily create your own.
Looking under the hood, we can notice that the program is written entirely in the Python programming language and uses the cross-platform Qt GUI toolkit for its graphical user interface, which means it runs on Linux, Microsoft Windows and Mac OS X operating systems.
The application is available for download as binary archives for many GNU/Linux operating systems, supporting both 32-bit (x86) and 64-bit (x86_64) instruction set architectures, as well as a source tarball.
What's new in relax 4.1.1:
- Mac OS X distribution file: Fixes for the DMG file generation. The .git directories are no longer bundled (the check in setup.py was for .svn directories), and the sobol_test.py script contained a bug that blocked the image generation.
- Changes:
- Release Checklist: Rewrite for the shift to a git repository and to the SourceForge infrastructure.
- Test suite: Temporary file fix for the Bmrb system and GUI tests. The temporary files normally used by these tests were accidentally removed in a previous commit. The result was temporary files being placed in the current directory.
relax 4.1.1
add to watchlist add to download basket send us an update REPORT- runs on:
- Linux
- main category:
- Science
- developer:
- visit homepage
IrfanView 4.67
ShareX 16.0.1
Zoom Client 6.0.3.37634
7-Zip 23.01 / 24.04 Beta
Windows Sandbox Launcher 1.0.0
calibre 7.9.0
Microsoft Teams 24060.3102.2733.5911 Home / 1.7.00.7956 Work
Context Menu Manager 3.3.3.1
4k Video Downloader 1.5.3.0080 Plus / 4.30.0.5655
Bitdefender Antivirus Free 27.0.35.146
- Context Menu Manager
- 4k Video Downloader
- Bitdefender Antivirus Free
- IrfanView
- ShareX
- Zoom Client
- 7-Zip
- Windows Sandbox Launcher
- calibre
- Microsoft Teams